Lab 3: SPARSITY-BASED LEARNING
This lab is about feature selection within the framework of sparsity based regularization, using elastic net regularization. Follow the instructions below. Think hard before you call the instructors!
Download file lab3.zip, extract it and add all the sub-folders to the path. - This file includes all the code you need!
Toy problem
We focus on a regression problem where the target function is linear. We will consider synthetic data generated (randomly sampled) according to a given probability distribution and affected by noise. You will have the possibility of controlling size of training and test sets, data dimension and number of relevant features.
Note that in the code we use a different notation from what you have seen in the classes. The functional minimized is:
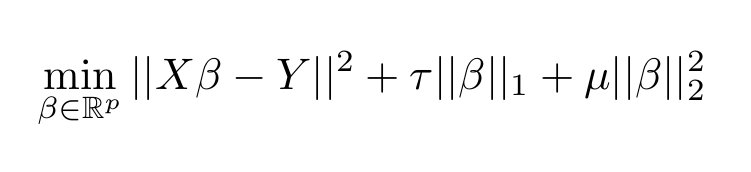
Overture: warm up
Run the file gui_l1l2 and the GUI will start.
Have
a look at the various components.
Generate a training set with the default parameters
press "run" to start a training phase with the selected L1_par and L2_par parameters and perform testing
change values for L1_par and L2_par and have a look at test error and number of selected variables;
first set L2_par=0 and vary L1_par trying to obtain a sparser or denser solution. What do you notice?
Repeat the experiment with a L2_par>0. How do test error and number of selected features vary?
now select KCV for L1_par tuning and observe the KCV error curve.
Interlude: the geek part
Back on the matlab shell, have a look to the content of directory "PROXIMAL_TOOLBOXES/L1L2_TOOLBOX". There you will find, among the others, the code for command "l1l2_algorithm" (used for variable selection), "l1l2_kcv" (used for model selection with kcv or loo), "l1l2_pred" (for prediction on a test set).
For more informations about the parameters and the usage of those scripts, type:
help l1l2_algorithm
help l1l2_kcv
help l1l2_pred
Finally,
you may want to have a look at file l1l2_demo_simple.m for a complete
example of analysis.
Allegro con brio: analysis
Carry out the following experiments either with the GUI, when it is possible, by personalizing the file demo_l1l2.m or by writing appropriate scripts.
(1)Prediction: Considering elastic net regularization, observe how the training and test error change
*when we change (increase or decrease) the regularization
parameter associated with the L1 norm
*when we change (increase or
decrease) the correlation parameter associated with the L2 norm
*
the training set size grows (try various choices of n in [10:....] as
long as matlab supports you!)
* the amount of noise on the
generated data grows (the test set is generated with the same
parameter of the training)
change one parameter at a time!
(2) Selection: Considering elastic net regularization, observe how the number and values of non zero coefficients in the solution change
*when we change (increase or decrease) the regularization
parameter associated with the L1 norm
*when we change (increase or
decrease) the correlation parameter associated with the L2 norm
*
the training set size grows (try various choices of n in [10:....] as
long as matlab supports you!)
* the amount of noise on the
generated data grows
(3) Large p and small n: Perform experiments similar to those above changing p (dimension of points), n (number of training points), s (number of relevant variables)
*set p<<n and s>n
*set p>>n and s>n
*set
p>>n and s<n
Crescendo: Data standardization (optional)
Data standardization: Consider the classification dataset given in part3-data.mat (use the scripts and NOT the gui):
*Use l1l2_algorithm to analyze the feature selected with different
values of the regularization parameters.
*Tune tau to select only
one variable, is there another variable that can provide a better
solution? (hint: only the first ten column of X are correlated with
Y)
*Can you figure out why the selected variable is not the one
that you would expect? (hint: analyse the correlation between the
columns of X and Y and the ranges of the columns of X, e.g. with
imagesc(X(:, 1:10)), colorbar)
Finale: Challenges
Challenge 1 - classification performances on microarray expression data
In the file part4-data.mat you can find a dataset of microarray
expression data. The original dataset is available here,
the given data has been extracted from the example data of L1L2
Signature.
The dataset contains 20 examples each of which
reports the expression levels of 7129 genes. The goal is to
distinguish between examples of acute lymphoblastic leukemia and
acute myeloid leukemia and to select the set of meaningful genes for
this task. The goal of this part is to maximize the precision of the
classification algorithm.
(1) Training:
Have
a look at the script "demo1_lab3.m". This script contains a
code snippet to perform feature selection using the previously
presented MATLAB scripts.
(2) Submit the results:
By the end of the challenge session - mandatory before the 6:00PM
- submit NAME_SURNAME-1 to http://www.dropitto.me/regmet (password:
regmet2013).
Challenge 2 - quality of the selected features on a function approximation problem
Consider the dataset in part5-data.mat. The dataset has been extracted from a generalized linear model. The goal of this part is to submit a complete list of functions that are correlated with the regression task.
(1) Training: Have
a look at the script "demo2_lab3.m". This script contains a
code snippet to perform feature selection using the previously
presented MATLAB scripts.
(2) Submit the results:
By the end of the challenge session - mandatory before the 6:00PM
- submit NAME_SURNAME-2 to http://www.dropitto.me/regmet (password:
regmet2013).